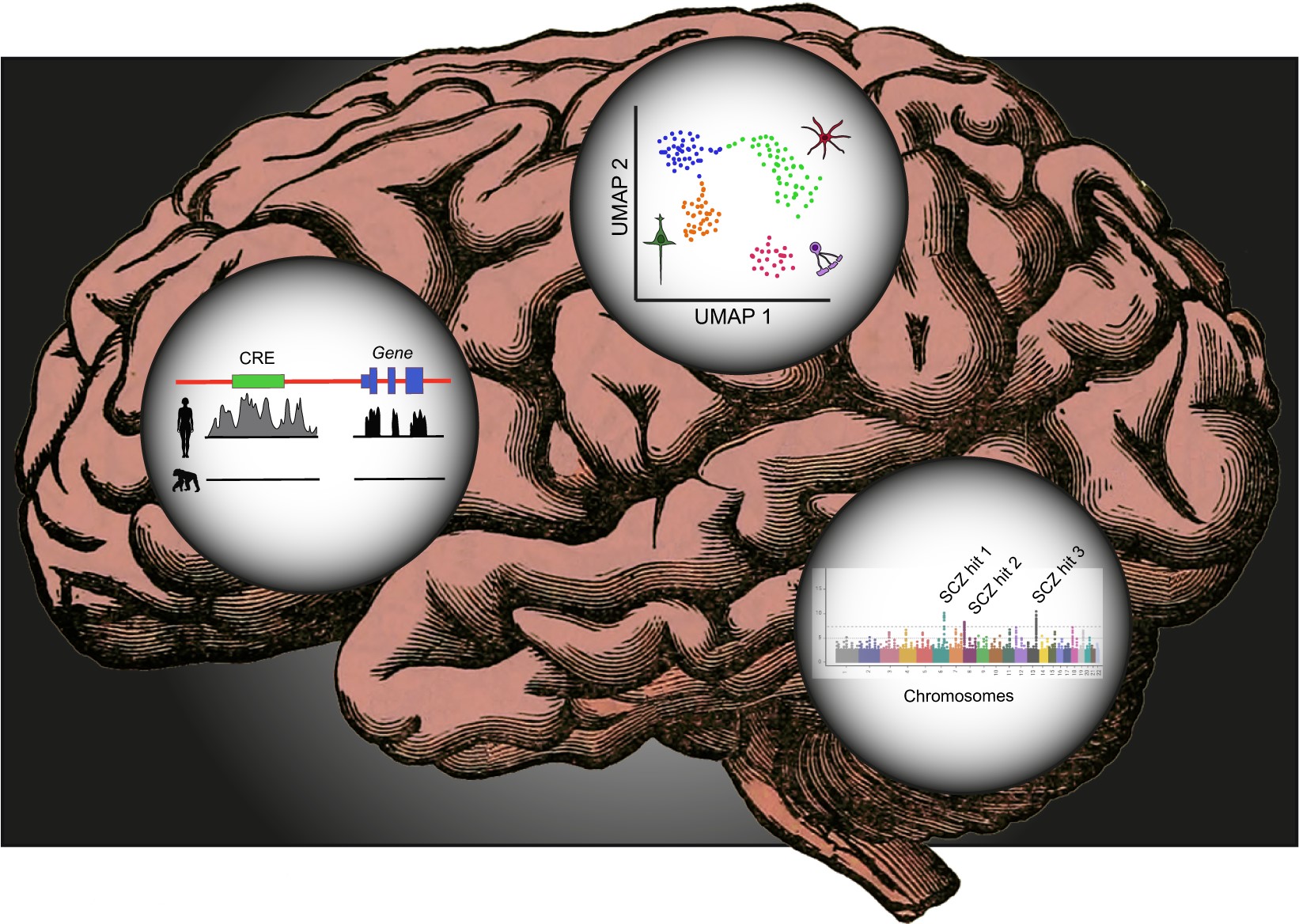
Research in the Neurogenomics group led by Gabriel Santpere aims to gain insight into the evolutionary mechanisms leading to human brain specializations by means of functional genomics. By leveraging multiple sources of OMICS data at the bulk tissue and single-cell/nuclei level, together with multiple sequence alignments, the group focuses on identifying lineage-specific variants relevant for normal primate brain development as well as for human neuropsychiatric disorders and traits.
Research lines
- Finding the needle in the haystack in the evolution of the human-specific features of brain development and neuropsychiatric traits
Humans possess a nervous system which confers very distinct cognitive abilities and very distinct cognitive disorders. The study of brain development is critical for the understanding of the evolution of these distinct features. To discern the genetic causes in evolution and disease influencing human-specific phenotypes, it is mandatory to identify the relevant variants affecting relevant genes among thousands of other variants predicted to be neutral. Even when a putative genetic-phenotypic connection has strong probability of causality, one still needs to demonstrate the mechanisms underlying the phenotypic change, which is an effort rarely pursued by evolutionary biologists. This line of research aims to deal with these two problems by i) reducing the search space for relevant variants into a tractable list, and ii) testing their functional effects in a system proximal to human fetal brain development consisting of iPSC-derived brain organoids.
- Genome-wide taxonomy of transcription factors binding sites across primates for the study of human brain specializations in health and disease
The study of the brain spatiotemporal convergence of risk for multiple neuropsychiatric traits has pointed to a reduced number of transcription factors with critical involvement in normal neurodevelopment. This line of research aims to dissect the temporal dynamics of genome-wide transcription factor binding site occupancy for a selection of risk convergence transcription factors (TF) at different stages of neurodevelopment and with an evolutionary perspective. This framework will provide the means to prioritize evolutionary relevant variants for human-specializations and to be tested in a CRISPR/Cas9 iPSC system for phenotypic characterization.
Website of group: https://www.imim.cat/programesrecerca/informaticabiomedica/neurogenomics.html